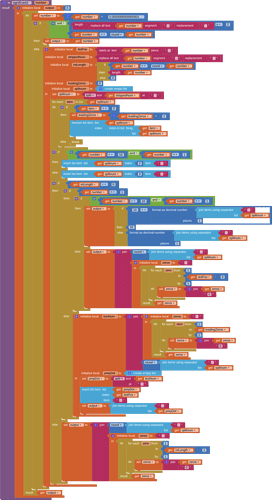
Hi, the code is working great, but occasionally I am getting numbers multiplied by 100, can't work out why. Do you mind taking a look? Thanks!
We are working without your code (.aia) and without any word on which values come out wrong, and without the base data.
That makes it hard to debug.
Sorry, have uploaded it now. Thanks, I really appreciate your help!
Cetacean_calculations_and_formulary.aia (57.1 KB)
Which values are coming out wrong?
You appear to divide most "weights" by 100, but some you do not, is this the issue ?
Here is an experiment you can try to see if the problem comes from the significant2 value procedure:
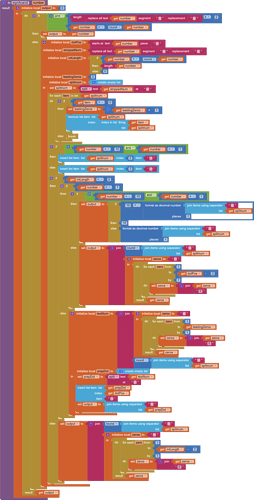
This is the procedure you currently use.
The idea is to replace it with another procedure that just returns its input, unchanged.
If that eliminates the 100 factor problem, the problem is in the procedure.
If it does not eliminate the 100 factor problem, the problem is in one or more of the calls to the procedure.
This is a delicate operation.
We can't change the procedure name or its parameter, or else all the call blocks will puke parameters all over the place.
Gently reach inside the procedure and yank out its local variable value block, leaving the procedure block empty but with its original name and parameter...
Plug the empty procedure block with the value of its input parameter...

Now run your tests.
To close up the patient after the tests, reverse the process...
remove the value block
re-insert the local variable block
I've tested it as you said, it was fine with the procedure just set to the input parameter, but when I put the full procedure back in, it produces the error again. It's not all the time, some of the results come out normal, and some will randomly seem to change, it seems to depend on what species and what weight I input on the previous screen. Different weights for the same species also through up the error as well.
If you want to proceed with the procedure you are currently using, help the author of that procedure by providing the numeric values that produce the problem.
Alternatively, use my procedure from
(pulling in both parts).
Hi, I have some examples:
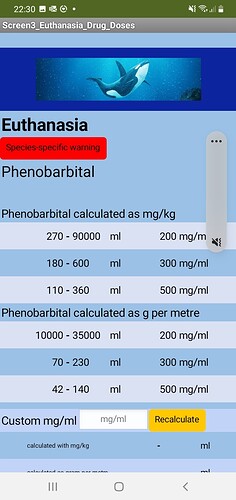
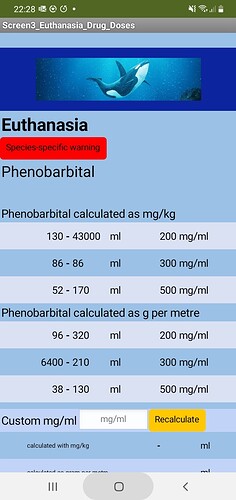
Harbour Porpoise (UK): 1.5m = 44kg is fine; 1.2m gives an error on the sixth calculation down on the right (phenobarb g per m 500mg/ml dose to).
Beluga whale: works fine on 3m = 365kg, fine on 4m, fine on 2.3m, but on 1.2m there is an error on the sixth calculation down on the right (phenobarb g per m 500mg/ml dose to).
Humpback whale: 17m = 70242kg error on fourth down on the left and right (phenobarb g per m 200mg/ml, dose from and to), and error on sixth calculation down on the left (phenobarb g per m 500mg/ml dose from). If 19m = 97520kg error on fifth calculation down on left (phenobarb 300ml/ml g per m dose from).
It seems very random, I really can't understand it.
Sorry. Do not know where to look in your blocks in order to see what calculations you are doing...
Please show the specific calculation for the displayed "incorrect" value returned.
I have tested all the numbers you show above, the procedure converts all of these correctly to two significant numbers
OK, believe I found the problem, when a number being added has a .0 at the end, e.g. 216.0, the procedure doesn't like it (wasn't a condition I expected/factored in)
Also, I get an alert from the app when calculating the weight for a beluga whale at 1.2m length. Comes back as 120kg, which can't be correct, if the weight for a harbour porpoise is 25kg?
Do you mind uploading the new block as a aia file? My computer won't allow the drag and drop png for some reason.
The lengths are mostly calculated with regression curves based on length and weight of previous animals, so a length too high or too small can give really odd numbers.
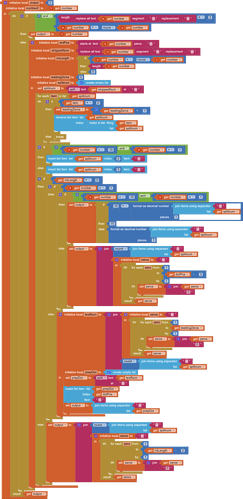
Just make and add this block to the existing procedure, first in the "do" block
(if you are likely to have any numbers with more that 9 decimal places, add more zeros!)
this seems to work perfectly! thanks so much!